
Research Activities
Research Activities
Publications
April 24, 2014
Study of the candidate factors responsible for the DNA demethylation required in iPS cell generation
A CiRA research team led by researcher Ren Shimamoto and senior lecturer Keisuke Okita has found that activation-induced cytidine deaminase (Aid), a candidate DNA demethylation factor, does not play an essential role in the iPS cell induction process. The research findings were published on April 9, 2014 (U.S. Eastern time) in the US scientific journal PLOS ONE.
It has been reported that, in the process of iPS cell induction, the cell's epigenetic status, specifically DNA methylation status and histone modification status, change from a somatic cell pattern to that of an ES cell. For instance, it is known that DNA in the promoter regions of Nanog and Oct3/4 and other pluripotent genes undergoes demethylation during iPS cell induction. It is also known that the DNA methylation level of these regions is higher in incompletely reprogrammed iPS cells than in iPS cells that have undergone complete reprogramming. DNA demethylation is therefore thought to play an important role in iPS cell induction, but the mechanism involved is not sufficiently understood.
One of the bases that constitute DNA is cytosine. Aid is a deaminating enzyme (deaminase), one of whose substrates is the methylated form of this cytosine. In recent years, it has been established that in early-stage zebrafish embryos, Aid contributes to DNA demethylation via the DNA repair pathway. It has also been reported to be involved in DNA demethylation in mouse early-stage embryo and brain. Additionally, when human fibroblasts are reprogrammed through fusion with mouse ES cells, Aid has been reported to be involved in the DNA demethylation of the promoter region of OCT3/4. These reports led the authors to hypothesize that Aid might also be connected to DNA demethylation during iPS cell generation. In order to test this theory, they generated iPS cells from Aid-knockout Nanog-GFP reporter mice1) and investigated the cells' properties.
When the four factors Oct4, Sox2, Klf4, and c-Myc were inserted into Aid-knockout mouse embryonic fibroblasts (MEF) and B cells via retroviral vectors, the iPS cell colonies obtained were GFP-positive like wild-type cells (Fig. 1). In the iPS cell induction rate, which indicates the number of GFP-positive colonies, no significant difference was found between wild-type and Aid-knockout cells. Inducing overexpression of Aid also had no effect on induction efficiency. These findings indicate that iPS cell generation efficiency was not influenced by whether Aid was present.
Next, the authors investigated the properties of Aid-knockout iPS cells. The proliferation ability of Aid-knockout iPS cells was similar to that of wild-type cells. A microarray was used to undertake a comprehensive analysis of intracellular gene expression, but only slight differences were found. It was also possible to create chimera mice using the Aid-knockout cells. These findings suggest that Aid-knockout iPS cells have equal self-replicating ability and differentiation potential to wild-type iPS cells.
To carry out a comprehensive analysis of the DNA-methylated regions of Aid-knockout iPS cells, MBD sequencing2) was performed. The results showed that the DNA-methylated regions of Aid-knockout and wild-type iPS cells were 99.5% identical, with only 0.5% difference. These findings suggest that the absence of Aid does not have a major influence on the DNA methylation status of iPS cells.
Taken together, the above results lead to the conclusion that Aid does not play an essential role in DNA demethylation during iPS cell generation.
The present study established that Aid does not play an essential part in DNA demethylation during iPS cell generation. This suggests the existence of a separate demethylation mechanism not connected with Aid. The study represents a valuable piece of research which should contribute to elucidating the mechanism of DNA demethylation during iPS cell generation.
1) Nanog-GFP reporter mouse
A gene-recombinant mouse model in which green fluorescent protein (GFP) is expressed together with Nanog expression. Nanog is known to be a gene with important actions in the maintenance of ES cell pluripotency and has been reported as a marker of iPS cell generation.
2) MBD sequencing
A method of detecting methylated regions which uses methyl-CpG binding domain (MBD) protein, which binds to methylated cytosine, to concentrate the methylated DNA before sequencing.
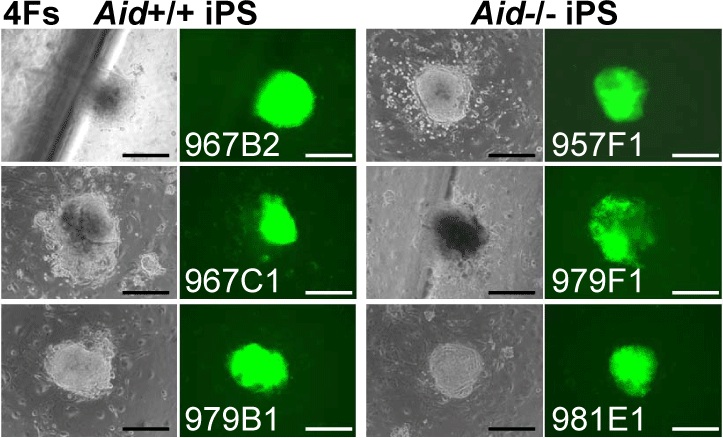
Fig. 1 iPS cell generation from Aid-knockout mice
Shape of Aid-knockout iPS cell colonies. The four factors were inserted into Aid-knockout MEFs and the colonies photographed after 25 days using a fluorescence microscope. Left-hand photos: phase contrast; right-hand photos: GFP fluorescence; bar: 200μm.
Paper Details
- Journal: PLOS ONE
- Title: Generation and Characterization of Induced Pluripotent Stem Cells from Aid-deficient Mice
- Authors: Ren Shimamoto, Naoki Amano, Tomoko Ichisaka, Akira Watanabe, Shinya Yamanaka, and Keisuke Okita






















