
News & Events
News & Events
News
August 04, 2015
RNA-based circuits to control cell behavior
Collaboration between the Massachusetts Institute of Technology (MIT) and the Center for iPS Cell Research and Application (CiRA), Kyoto University, has led to the invention of RNA-based gene circuits that control cellular output. Because these circuits are entirely RNA-based, they should be safer to use in humans than their DNA-based counterparts and therefore available for a number of biomedical applications.
A common way to control cell function is through the use of drugs or other small compounds. However, drug actions are often imprecise and risk undesirable side effects. A better tool would be synthetic gene circuits. Unfortunately, although a number of such circuits already exist, none are suitable for biomedical applications and therefore cannot replace drugs. "All circuit designs rely exclusively or partially on DNA-based transcriptional regulation, and the required DNA poses a risk of cancer," says Ron Weiss, a synthetic biologist at the MIT who is seeking ways around this problem.
Accordingly, Weiss considered ways to remove the requirement for transcriptional regulation by making RNA-only circuits. To push this work quickly, he partnered with Hirohide Saito, a bioengineer and professor at CiRA who specializes in RNA-based technologies. Saito explains what makes the RNA-only circuits envisaged by Weiss more challenging. "In the case of DNA, various transcriptional repressors and activators are known and used to construct circuits. In the case of RNA-only circuits, we need a set of RNA-binding proteins that effectively control gene expression in a post-transcriptional manner."
One candidate protein is the archaeal protein L7Ae, which selectively binds kink-turn RNA and therefore can be used in synthetic circuits to inhibit the expression of another protein. As a first example, the scientists prepared two synthetic RNA, one that included mRNA for the expression of L7Ae and the other mRNA for the expression of hBax, a pro-apoptosis protein that when expressed kills the cell.
Along with these mRNA, included in the synthetic RNA were antisense sequences for micro RNA (miRNA). Binding of miRNA to the synthetic RNA prevents the expression of the mRNA. Therefore, if an undesired cell type has unique miRNA, one could design the circuit to include antisense sequences for this miRNA to prevent the expression of L7Ae, while antisense sequences for miRNA unique to a desired cell type could be added so as to prevent the expression of hBax.
While miRNA alone can be sufficient to separate two cell types, Weiss and Saito show that their RNA circuits can separate one cell type from multiple cell types simultaneously. This ability is important for advancing cell therapies, as these require first acquiring the desired cell type from a heterogeneous pool of cells before being transplanted into a patient.
Because the synthetic RNA is not needed once the cells are sorted, they should only be transiently expressed. For this reason, modified mRNA (modRNA) was used to deliver the synthetic RNA into the cells, since it has a short half-life. In contrast, when a more permanent effect on the cell is sought, such as regulation of a signaling cascade, an alternative vehicle is needed, such as RNA replicons (replicons).
To extend the application of their circuits, replicons were also tested as a delivery system and found effective, as they could turn the expression of multiple proteins off and on. Importantly, the circuit was easy to scale, which in theory should allow this technology to control even more targets at any given time.
The ability to construct RNA-only circuits and deliver them using different modes has made Weiss and Saito optimistic about the wide applicability of their circuits and its potential for use in humans. "The ability to use modRNA and replicons greatly expands the reach of our circuits", said Saito.
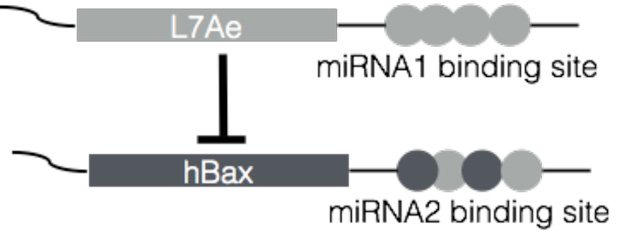
RNA-based circuits can separate a desired cell type from multiple cell types simultaneously. In this illustrative example, the expression of L7Ae will repress the expression of hBax, allowing the cells to survive. If, however, miRNA1 binds to its binding site, L7Ae expression is repressed, which allows hBax to be expressed, killing the cell. If miRNA2 binds to its binding site, hBax is expressed, regardless of L7Ae.
Paper Details
- Journal: Nature Biotechnology
- Title: Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery
- Authors: Liliana Wroblewska, Tasuku Kitada, Kei Endo, Velia Siciliano, Breanna Stillo, Hirohide Saito, Ron Weiss






















