
News & Events
News & Events
News
February 19, 2024
Combining expertise to characterize the SARS-CoV-2 Omicron XBB.1.5 variant
The respiratory virus SARS-CoV-2, responsible for the COVID-19 pandemic, continues to evolve. A better understanding of how it is evolving will be crucial when considering our current defenses (e.g., vaccines) against it and guiding our preparations for future viral pandemics.
By reconstructing the phylogenetic trees of the XBB lineage, the researchers verified that XBB.1.5 most likely emerged from an XBB.1 ancestral strain. From this analysis, they also concluded that while the S486P mutation in the spike (S) protein had arisen as the XBB.1.5 sublineage diverged from other XBB.1 variants, the G252V mutation in the S protein and the truncating mutation in ORF8 (G8stop) occurred during the diversification of the XBB.1 lineage. These findings suggest that the S486P mutation likely contributed to the enhanced fitness of the XBB.1.5 variant.
Importantly, by examining whether XBB.1.5 is resistant to sera collected from donors following vaccination with various SARS-CoV-2 vaccines, the researchers determined the XBB.1.5 variant to possess similar immune evasiveness as the XBB.1 parent strain. Similarly, XBB.1.5 S protein was observed to have comparable membrane fusion ability as that of XBB.1. The research team also examined the structural properties of the S protein via cryogenic electron microscopy to understand the structural basis of its enhanced immune evasiveness and determine the effects of the S486P mutation on its interaction with the ACE2 receptor, the primary cell entry mechanism exploited by the SARS-CoV-2 virus. While some local structural differences were detected between the S proteins of XBB.1 and XBB.1.5, they display high overall structural similarity, consistent with the comparable immune evasiveness and membrane fusion ability observed.
The researchers next performed in vitro infection experiments on cultured cells, in which they observed slower growth for XBB.1 and XBB.1.5 compared to the Delta variant but also noticed a slightly higher growth rate for XBB.1.5 compared to XBB.1. Morphological analysis of the infected cells revealed while infection by the Delta variant resulted in large multinuclear syncytia, XBB.1- and XBB.1.5-infected cells showed smaller syncytia. Notably, cells infected by XBB.1.5 possessed an intermediate number of syncytia, consistent with a stronger binding affinity to ACE2, as reported previously. In addition, the research team also infected airway-on-a-chip, an experimental setup to recapitulate the airway epithelial-endothelial barrier, and found that both XBB.1 and XBB.1.5 have reduced ability to disrupt this barrier compared to the Delta variant.
In addition, the research team examined viral infections in a hamster model to better understand its in vivo viral pathogenicity. In these experiments, animals infected with XBB.1.5 experienced less weight loss compared to those infected with either XBB.1 or the Delta variant, and while similar viral loads were detected in respiratory tissues, bronchitis or bronchiolitis caused by XBB.1.5 was less severe compared to XBB.1.
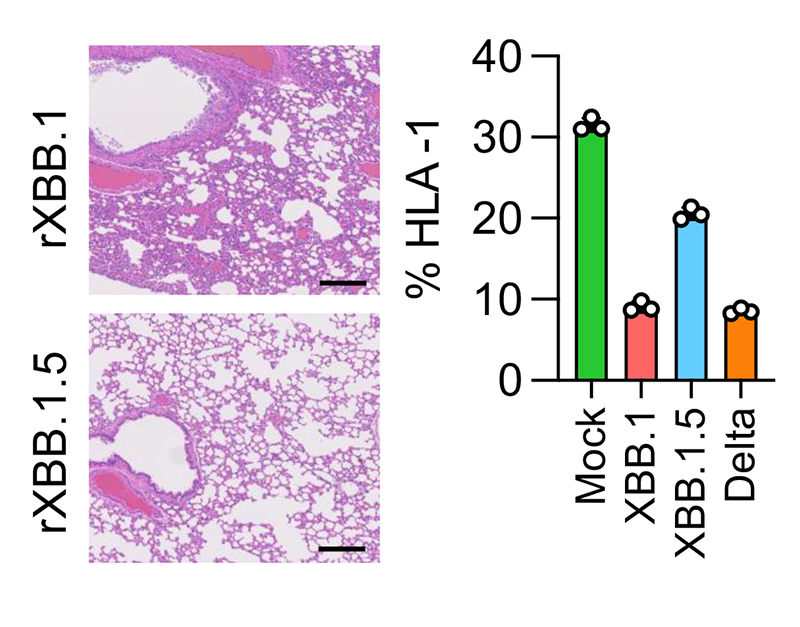
Finally, the effects of the S protein S486P mutation (S:S486P) and the G8Stop mutation in ORF8 (ORF8:G8Stop) were examined by incorporating them separately into the parental XBB.1 backbone for direct comparison with XBB.1 and XBB.1.5. In vivo hamster infection experiments using these viral strains revealed XBB.1 S:S486P to cause greater body weight loss and more severe bronchitis like XBB.1, whereas XBB.1 ORF8:G8Stop led to milder effects that were comparable to XBB.1.5. Using human iPS cell-derived lung organoids, the researchers also observed a milder decline in MHC-I expression caused by XBB.1.5, in comparison to XBB.1 and the Delta variant, consistent with a previous report suggesting ORF8 encodes a cytokine responsible for suppressing MHC-I expression. Furthermore, histological analysis revealed reduced inflammation in hamster respiratory tissues infected with XBB.1 possessing only the ORF8:G8Stop mutation.
Together, through a comprehensive analysis conducted by the G2P-Japan Consortium, both S:S486P and ORF8:G8Stop mutations unique to XBB.1.5 are involved in the pathogenicity displayed by this variant. Notably, this Omicron subvariant has acquired mutations that enhance viral spread while diminishing its pathogenic potential. While it remains to be seen how continued evolution will affect virological characteristics, collaborative efforts such as those displayed in this study provide hope that we are more equipped to study SARS-CoV-2 in detail than ever before.
Paper Details
- Journal: Nature Communications
- Title: Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant
- Authors:
Tomokazu Tamura1,2,3,4,5,**, Takashi Irie6,**, Sayaka Deguchi7,**, Hisano Yajima8,**, Masumi Tsuda2,3,9,**, Hesham Nasser10,**, Keita Mizuma11,**, Arnon Plianchaisuk12,**, Saori Suzuki1,2,3,4,**, Keiya Uriu12, M. S. T, Monira Begum10, Ryo Shimizu10, Michael Jonathan10, Rigel Suzuki1,2,3,4, Takashi Kondo3, Hayato Ito2, Akifumi Kamiyama2, Kumiko Yoshimatsu13, Maya Shofa14, Rina Hashimoto7, Yuki Anraku15, Kanako Terakado Kimura8, Shunsuke Kita15, Jiei Sasaki8, Kaori Sasaki-Tabata16, Katsumi Maenaka15,16, Naganori Nao5,11, Lei Wang2,3,9, Yoshitaka Oda1,2,3, The Genotype to Phenotype Japan (G2P-Japan) Consortium, Terumasa Ikeda10, Akatsuki Saito14, Keita Matsuno5,11, Jumpei Ito12, Shinya Tanaka2,3,9,*, Kei Sato10,12,*, Takao Hashiguchi8,*, Kazuo Takayama7,*, Takasuke Fukuhara1,2,3,4,5,17,*
**: These authors contributed equally
*: Corresponding authors - Author Affiliations:
- Faculty of Medicine, Hokkaido University
- Graduate School of Medicine, Hokkaido University
- School of Medicine, Hokkaido University
- Institute for Vaccine Research and Development (IVReD), Hokkaido University
- One Health Research Center, Hokkaido University
- Graduate School of Biomedical and Health Sciences, Hiroshima University
- Center for iPS Cell Research and Application (CiRA), Kyoto University
- Institute for Life and Medical Sciences, Kyoto University
- Institute for Chemical Reaction Design and Discovery (WPI-ICReDD), Hokkaido University
- Joint Research Center for Human Retrovirus Infection, Kumamoto University
- International Institute for Zoonosis Control, Hokkaido University
- The Institute of Medical Science, The University of Tokyo
- Institute for Genetic Medicine, Hokkaido University
- Department of Veterinary Science, Faculty of Agriculture, University of Miyazaki
- Faculty of Pharmaceutical Sciences, Hokkaido University
- Graduate School of Pharmaceutical Sciences, Kyushu University
- Research Institute for Microbial Diseases, Osaka






















