
News & Events
News & Events
News
September 06, 2024
Leveraging the power of iPS cell technology to study myeloid neoplasm
There are various causes for cancers, but a primary means for their development is the rearrangement of chromosomes that allows cancer-causing genes to hijack enhancer elements from other genes to increase their expression aberrantly. For example, recurrent rearrangements on chromosome 3 (inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2)) place a GATA2 enhancer close to the MECOM (EVI1) promoter to drive hematological malignancies by aberrantly enhancing MECOM gene expression. More recently, a specific type of leukemias, known as myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML) with atypical 3q26 rearrangements including t(3;8)(q26.2;q24), in which chromosomal changes also result in abnormally high expression of MECOM was characterized. While these chromosomal rearrangements are rare, the resulting cancer has a poor prognosis with a median survival of only about 6 months and, as such, was designated as a new category of AML with MECOM rearrangement by the World Health Organization in 2022.
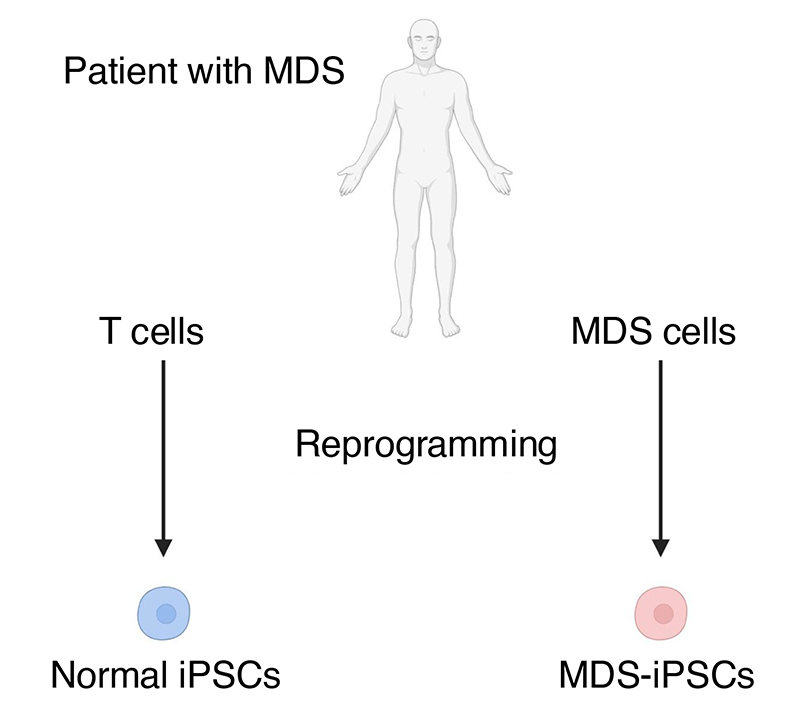
To gain insights into this newly categorized disease and identify potential therapeutic strategies, the research team successfully established iPS cells from an MDS patient with t(3;8)(q26.2;q24) (MDS-iPSCs), which showed the same chromosomal abnormalities as cells originally collected from the patient. Specifically, chromosomal breakpoints were identified upstream of the MECOM (MDS1-EVI1 and EVI1) promoters on 3q26.2 and upstream of the MYC super-enhancer on 8q24. They subsequently differentiated these MDS-iPSCs into hematopoietic progenitor cells (MDS-HPCs) as an MDS disease model. As expected, MDS-HPCs displayed leukemic morphology and showed defects for differentiating into myeloid (granulocytes and monocytes) and erythroid (red blood cells) lineages.
Consistently, gene expression analysis by RNA sequencing revealed many dysregulated genes, with reduced expression of numerous factors critical for hematopoietic differentiation and, conversely, the expression of many regulators crucial to HSC maintenance enhanced in MDS-HPCs compared to HPCs generated from isogenic normal iPS cells. Notably, MDS-HPCs displayed an AML-like gene expression signature. Furthermore, MECOM gene expression was increased dramatically in MDS-HPCs, consistent with the hypothesized causal role in the disease. Epigenetic analyses further indicated that the activity of the MDS1-EVI1 and EVI1 promoters was markedly increased in MDS-HPCs, consistent with enhanced MECOM expression in this patient's HPCs due to the translocation of the MYC super-enhancer.
Because BRD4, a member of the bromodomain and extraterminal (BET) protein family, accumulates at super-enhancers, the researchers speculated that its inhibition could suppress the expression of cancer-causing genes. To test this hypothesis, they treated MDS-HPCs with JQ1, a BRD4 inhibitor, and monitored MECOM expression. While JQ1 treatment did not affect control HPCs, it significantly lowered MECOM expression in MDS-HPCs. Consistent with this, a fluorescent reported-based assay revealed that JQ1 effectively reduces gene expression from the EVI1 locus. Most importantly, however, JQ1 treatment led to increased cell death of MDS-HPCs in a dose-dependent manner but did not affect control HPCs, thus demonstrating its therapeutic potential against MDS/AML.
Altogether, this work demonstrates again the power of iPS cell technology for modeling human diseases and reinforces it as an effective means of testing promising therapies that may be useful in clinics as treatments and cures.
Paper Details
- Journal: British Journal of Haematology
- Title: Modelling and drug targeting of a myeloid neoplasm with atypical 3q26/MECOM rearrangement using patient-specific iPSCs
- Authors:
Momoko Nakamura1,2, Kazuhisa Chonabayashi1,2*, Megumi Narita1, Yasuko Matsumura1, Misato Nishikawa1, Yotaro Ochi3, Yasuhito Nannya3,4, Masakatsu Hishizawa2,5, Daichi Inoue6, Ruud Delwel7,8, Seishi Ogawa3, Akifumi Takaori-Kondo2, and Yoshinori Yoshida1*
*:Corresponding authors - Author Affiliations:
- Center for iPS Cell Research and Application, Kyoto University
- Department of Hematology, Graduate School of Medicine, Kyoto University
- Department of Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University
- Division of Hematopoietic Disease Control, The Institute of Medical Science, University of Tokyo
- Department of Hematology, Kyoto Katsura Hospital
- Department of Hematology-Oncology, Institute of Biomedical Research and Innovation, Foundation for Biomedical Research and Innovation at Kobe
- Department of Hematology, Erasmus MC Cancer Institute
- Oncode Institute






















