Research Activities
Research Activities
Home › Research Activities › Publications › Three-dimensional design method for protein-responsive RNA switch to regulate cell fate
Publications
January 18, 2013
Three-dimensional design method for protein-responsive RNA switch to regulate cell fate
In a joint research project with Professor Tan Inoue (Kyoto University Graduate School of Biostudies), the research group of Associate Professor Hirohide Saito and Shunichi Kashida (CiRA Department of Reprogramming Science) has used three-dimensional modeling to perform molecular design of RNA-protein interactions, resulting in the successful development of an RNA switch(1) capable of regulating the effect of RNA interference (RNAi)(2) in response to intracellular conditions. The research findings were published in the British scientific journal Nucleic Acids Research on October 18, 2012, as a Featured Article - a category representing the top 5% of articles in terms of quality - and appeared on the journal's front cover.
By modifying short-hairpin RNA (shRNA)(3), the research group had previously succeeded in developing an RNA switch to regulate the survival of target cells expressing specific proteins (Saito H., et al. Nat. Commun. 2:160, 2011). This switch was able to regulate target gene expression in response to intracellular production of specific proteins. However, before this device could be applied in medical or biological science, including in the differentiation of iPS cells into target cell types, researchers still needed to establish the relevant action mechanism so as to develop switches responsive to a range of human-derived proteins.
shRNA causes the phenomenon known as RNA interference (RNAi), whereby target gene expression is suppressed through cleavage by Dicer (an RNA-cleaving enzyme). If a specific protein binds RNA in such a way as to block the Dicer cleavage site, it can therefore be expected to prevent RNA interference. The researchers first of all used 3D molecular modeling software to create a computer-based reproduction of the 3D structure of the shRNA switch and Dicer and of their spatial relationship at the time of cleavage. This indicated that altering the length of the shRNA double-chain site might make it possible to adjust the position of the protein that binds to the Dicer cleavage site in such a way as to bring it closer toward collision with Dicer.
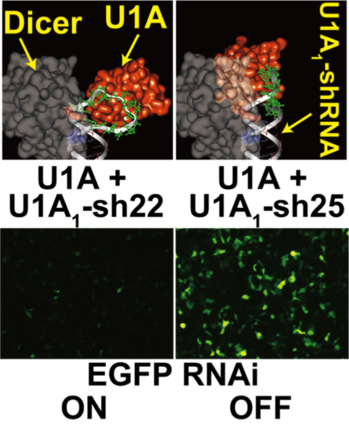
In the next step, the intracellular introduction of an shRNA switch that inhibits cleavage by Dicer through binding of U1A (a protein present in human cells) did indeed show that the action of RNAi could be suppressed in response to intracellular U1A production (Figure 1). Also, to confirm the versatility of the 3D molecular design method, an RNA sequence that binds to a transcription factor (NF-kB) expressed in a range of cancer cells was incorporated in the shRNA. As expected, it was found that the effect of RNAi could be suppressed in response to the intracellular expression of this transcription factor.
Figure 1 3D molecular model showing: above: projected 3D steric hindrance between Dicer and the protein binding to the shRNA switch; below: evaluation of the intracellular function of the shRNA switch
With an shRNA predicted not to inhibit the action of Dicer (above left), RNAi against enhanced green fluorescent protein (EGFP) was not suppressed and no expression of EGFP was observed (below left). With the shRNA predicted to inhibit cleavage by Dicer (above right), RNAi was blocked and EGFP expression was observed (below right).
The researchers applied 3D molecular modeling to the molecular design of RNA-protein interactions to successfully develop an RNA switch capable of regulating the effect of RNA interference (RNAi) in response to the presence or absence in human cultured cells of specific proteins. This design method can be used to predict and to optimize the intracellular function of RNA switches responsive to specific proteins. Among the future applications of this method will be technology to regulate the fate of target cells in response to intracellular conditions and technology to efficiently select only cells that have differentiated into a specific cell type. It is thus envisaged as a new tool for developing methods of inducing differentiation of iPS cells into target cells.
The present research was carried out in partnership between Kyoto University Center for iPS Cell Research and Application, Kyoto University Graduate School of Biostudies, and the Kyoto University Hakubi Project.
Title of Paper
Authors
Shunnichi Kashida, Tan Inoue*, and Hirohide Saito*
* corresponding author
Funding
The present research was undertaken with financial support from the following institutions
1. Japan Science and Technology Agency, International Cooperative Research Project (ICORP), RNA Synthetic Biology Project
2. New Energy and Industrial Technology Development Organization, Financial support to young researchers
3. Ministry of Education, Culture, Sports, Science and Technology, Grants-in-Aid for Scientific Research; Young Scientists A
4. Japan Society for the Promotion of Science, Research Fellowships for Young Scientists (DC2)
5. Takeda Science Foundation, Life Science Research Grant
Notes
1) RNA switch
The present research used RNA-protein interactions triggered by shRNAs that induce RNAi to render the RNAi mechanism inoperative only in the case of intracellular expression of the target protein. In other words, when target protein A is not expressed, the RNAi mechanism operates so that the target mRNA is cleaved and synthesis of protein B from the target mRNA is switched off. However, when target protein A is expressed, the mechanism of RNAi is inhibited, the target mRNA is not cleaved, and the synthesis of protein B proceeds.
2) RNA interference (RNAi)
A method of suppressing the expression of a chosen gene using double-chain RNA formed within the cell. Its originators, Andrew Fire and Craig Melo, were awarded the 2006 Nobel Prize for Physiology or Medicine. Research is now progressing into the application of RNAi to drug research.
3) shRNA
Abbreviation of short hairpin RNA, a kind of RNA named for its shape. It is used to suppress the action of RNAi on target genes.