
Research Activities
Research Activities
Publications
March 08, 2019
Gene editing iPS cells simplifies patient-donor matching
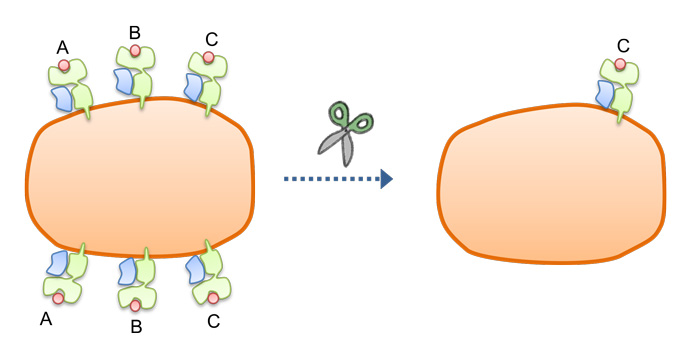
The knockout of all but one HLA allele by gene editing simplifies donor-patient matching with while minimizing the risk of an immune reaction.
The clinical application of iPS cells has already began, with patients receiving transplantations of cells made from iPS cells to treat eye and brain diseases. However, the number of patients who can benefit from these and future therapies is limited due to donor-patient matching. In a new study seen in Cell Stem Cell, the Akitsu Hotta and Shin Kaneko laboratories report a gene-editing strategy could bring iPS cell therapies to a wider range of patients.
One of the greatest limitations in cell therapies is immune matching. The transplantation of cells from a mismatching donor could lead to complications more serious than the disease itself, and even death, all because the immune system of the patient reacts aggressively against what it perceives as a pathogenic invasion. Donors who share the same human leukocyte antigens (HLA) as the patient are said to match and therefore do not elicit a dangerous immune response. To maximize the number of patients who will benefit from iPS cell-based therapies, CiRA is recruiting donors who are HLA homozygous, as these donors match with a much larger number of the general population than those who are HLA heterozygous. However, donors who are HLA homozygous are rare.
"140 cell lines are needed to cover >90% of the Japanese population," explains Hotta. "Each cell line comes from a unique HLA homozygous donor, and to find 140 donors homozygous for HLA-A, -B, and -DR, it is estimated that approximately 160,000 people will have to be tested."
To reduce the number of people tested, several scientists have suggested that rather than recruiting HLA homozygous donors, gene editing the HLA of any donor to homozygosity may be more efficient. HLA has many varieties, but the ones most essential for immune matching involve HLA class I, which include the subsets HLA-A, HLA-B, and HLA-C, and HLA class II, which include the subsets HLA-DP, HLA-DQ, and HLA-DR.
"Mismatching in HLA class I activates CD8 killer T cells, and mismatching in HLA class II activates CD4 helper T cells," explains Dr. Huaigeng Xu, the first author the study. "If there is no HLA expression, then there is no T cell response."
One logical approach then would be to use gene editing technology to remove all HLA class I and II expression. However, minus this expression, the immune system may not respond to infecting pathogens. At the same time, it will respond to HLA-null cells through natural killer (NK) cells. "Our solution was to make pseudo-HLA homozygous iPS cells," continues Xu.
Each HLA subset is expressed two alleles. HLA homozygous means that the two alleles are the same. So long the patient has one HLA allele that is the same as the two homozygous HLA alleles of the donor, then there is a match. Xu proposed using gene editing to erase one HLA allele from potential donor iPS cells.
He hypothesized that matching would be successful so long the patient had one HLA allele that was the same as the remaining HLA on the donor. Indeed, experiments showed exactly this effect. Matching patients with pseudo-homozygous HLA class I donors evade CD8 killer T cell responses.
However, even this strategy would require a large number of lines to serve the whole Japanese population. Therefore, Xu considered more gene editing.
"We deleted both HLA-A, HLA-B and mono HLA-C alleles to retain one HLA-C allele. We called these cells HLA-C retained cells," he says. Like pseudo-homozygous HLA class I, these cells evaded CD 8 T cells, and also evaded NK cells. More importantly, fewer lines are needed to serve a large population.
Finally, to also evade CD4 helper T cells, the group deleted HLA class II from the HLA-C retained cells. These cells evaded CD8 killer T cells, CD4 helper T cells, and NK cells. The research group estimates that a core stock of 12 HLA-C retained iPS cell lines could serve the multiple populations.
Paper Details
- Journal: Cell Stem Cell
- Title: Targeted Disruption of HLA genes via CRISPR-Cas9 generates iPSCs with Enhanced Immune Compatibility
- Authors: Huaigeng Xu1*, Bo Wang1*, Miyuki Ono1,2, Akihiro Kagita1, Kaho Fujii1, Noriko Sasakawa1,2, Tatsuki Ueda1, Peter Gee1,2, Misato Nishikawa1, Masaki Nomura1, Fumiyo Kitaoka1, Tomoko Takahashi1, Keisuke Okita1, Yoshinori Yoshida1, Shin Kaneko1#, and Akitsu Hotta1,2#
* co-first authors
# co-corresponding authors - Author Affiliations:
- Center for iPS Cell Research and Application (CiRA), Kyoto University, Kyoto, Japan
- Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development (AMED), Tokyo, Japan






















