
Research Activities
Research Activities
Publications
October 18, 2013
Global Splicing Pattern Reversion during Somatic Cell Reprogramming
A research team led by Takuya Yamamoto, assistant professor at CiRA who works for iCeMS, and postgraduate CiRA researcher Shou Ohta at Kyoto University Graduate School of Biostudies and has established that, in the process of reprogramming from somatic cells to iPS cells, the splicing pattern which translates the RNA is also subject to reprogramming. This finding has been published in Cell Reports on October 18.
Alternative splicing is one of the systems that allows a number of proteins to be produced from a single gene, making possible the production of a richer range of protein types and thereby enabling more complex and flexible responses. Differentiated cells all have a specific splicing pattern which produces the functions and characteristics peculiar to each cell type.
Many diseases reportedly arise from disturbances in the splicing pattern. It has also been reported that there is a splicing pattern specific to ES cells, which and have unlimited ability to proliferate and are pluripotent, which means that ES cells are capable of differentiating into any cell type). In other words, the splicing pattern can be seen as a major factor in determining cell characteristics.
Differentiated cells can be reprogrammed into iPS cells by introducing reprogramming factors, but it had remained unclear whether the splicing pattern is also modified in this process. If no change occurred in the splicing pattern, the same group of iPS cells might display major differences in their properties depending on the cell type they were derived from. To clarify this area, Yamamoto and his fellow researchers used large-scale gene analysis technology to examine the splicing patterns of somatic cells and iPS/ES cells.
They found that the splicing pattern of the somatic cells had reverted to that of a pluripotent cell (Fig. 1). Specifically, the splicing pattern of the iPS/ES cells was closely similar to that of gonad cells.
The researchers further established that the process for creating cells with a pluripotent splicing pattern involves the regulation of splicing by RNA binding proteins that act specifically on iPS/ES cells and produce the characteristic pattern. The researchers found that when two of these RNA binding proteins, U2af1 and Srsf3, were deactivated, the success rate of conversion from somatic cell to iPS cell was reduced, indicating that U2af1 and Srsf3 play an important role in reprogramming.
The researchers analyzed alternative splicing across the whole of the genome and discovered that the mechanism that controls the splicing patterns and splicing in the cell reprogramming process is subject to modification. The findings also suggest that the control of alternative splicing is one of the major inputs in the cell reprogramming mechanism and has an important influence on pluripotency.
The findings show that, in iPS cells, in the same way as in ES and other cell types, the splicing pattern changes to a pluripotent pattern during conversion from somatic cells. Application of these findings should assist in the evaluation of iPS cell quality and help improve for instance the success rate of iPS cell generation and shorten the time required.
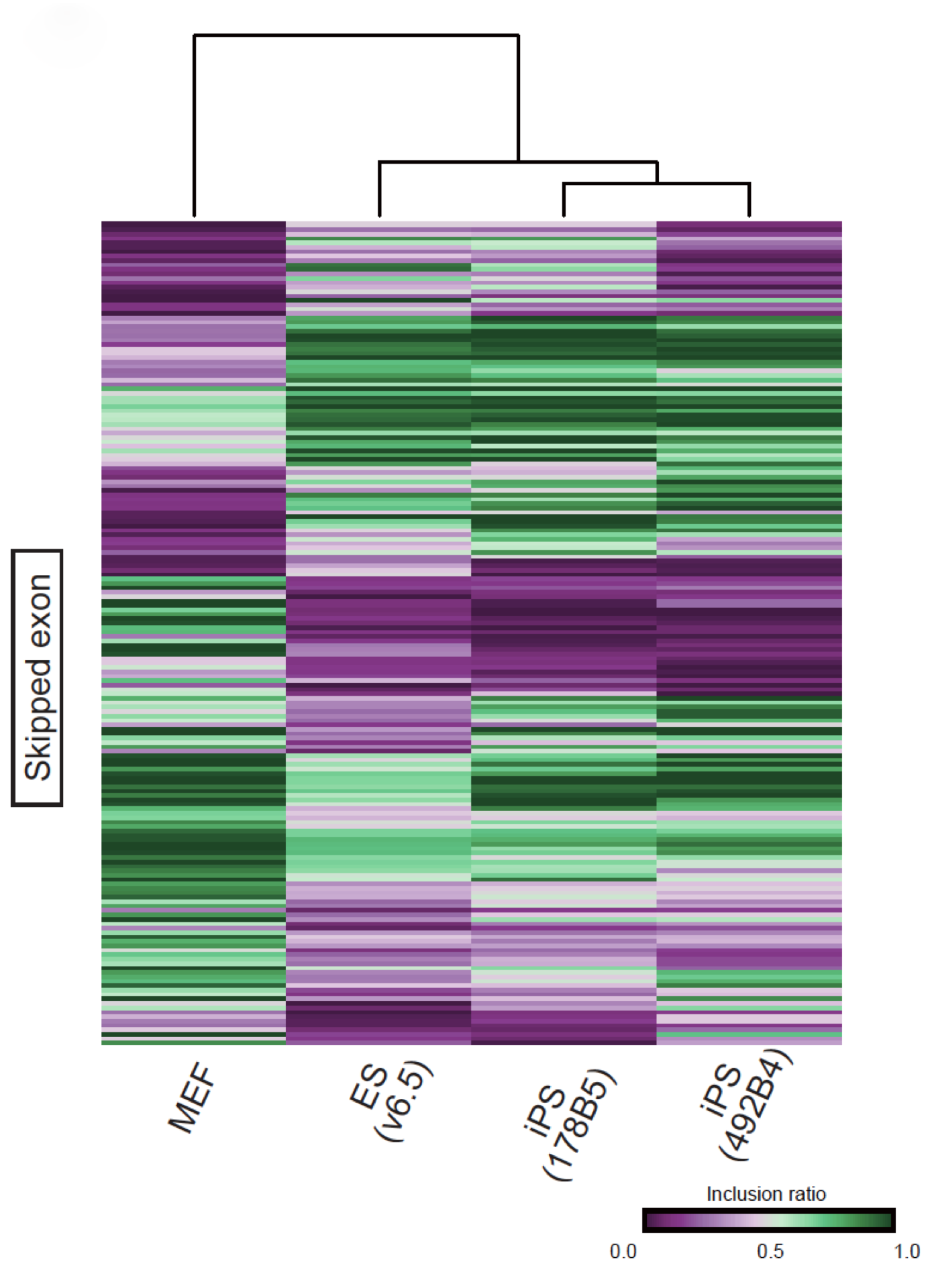
Fig. 1 Analysis of splicing pattern
There are various different types of alternative splicing; the figure here shows an analysis of skipped exons (the choice between A-B-C and A-C, i.e. whether exon B is skipped). iPS cells generated from mouse embryonic fibroblasts (MEF) were found to differ greatly from MEF in having a splicing pattern similar to that of ES cells.
<Journal Information>
Title of Paper
Global Splicing Pattern Reversion during Somatic Cell Reprogramming
Authors
Sho Ohta, Eisuke Nishida, Shinya Yamanaka and Takuya Yamamoto






















