
Research Activities
Research Activities
Publications
January 11, 2019
OVOL1 puts the brakes on partial reprogramming
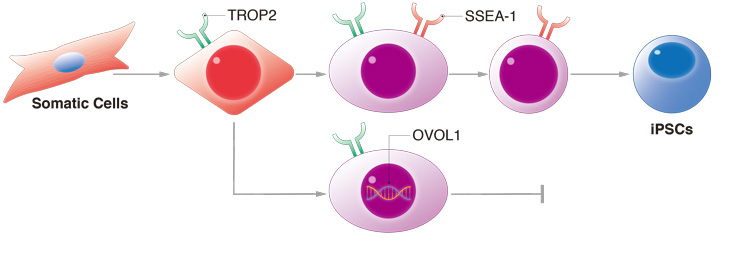
The transient expression of TROP2 was found to have a high correlation with iPSC reprogramming, while the expression of OVOL1 was found to repress the growth of cells that failed to reprogram.
A new study by the laboratory of CiRA Associate Professor Knut Woltjen has revealed two new factors that contribute to the reprogramming of cells into iPS cells. The transcription factor OVOL1 was found to control the growth of cells that do not proceed along the fate of iPS cells, and the surface protein TROP2 was found to identify the cells. The discovery, which can be read in Stem Cell Reports, is expected to improve the efficiency of iPS cell generation, providing new reprogramming strategies that could lower costs for experimental cell therapies and drug discovery.
The original and standard reprogramming method to create iPS cells requires four genes - OCT3/4, SOX2, KLF4, and c-MYC - also known as the "Yamanaka factors." The Woltjen laboratory had found previously that the relative amounts, or stoichiometry, of the Yamanaka factors influences the probability of iPS cell generation.
"We found using more KLF4 drastically improved the reprogramming quality at the cost of yield," says Woltjen, "but couldn't explain why."
Reprogramming begins with cells losing their identity. Once their identities are lost, they can proceed along a pathway towards iPS cells. However, while many cells can begin to reprogram, the majority fail to complete the process. These partially reprogrammed cells take on other identities which do not share the pluripotency of iPS cells and thus cannot be used for studying human development or in medical applications.
Following up on their discovery of KLF4 stoichiometry, doctoral student Harunobu Kagawa realized that cells reprogrammed with more KLF4 had distinct protein signatures on their surface.
"Most cells that finished reprogramming to become iPS cells had expressed TROP2 transiently before expressing SSEA-1," he says.
SSEA-1 is recognized as a key marker for reprogramming to iPS cells and pluripotency. Kagawa found that most cells successfully reprogrammed with more KLF4 would express TROP2 before SSEA-1, and would stop expressing TROP2 shortly thereafter. Cells that kept expressing TROP2 remained in a partially reprogrammed state.
"Most cells that failed to reprogram also failed to remove the TROP2 mark," he notes.
Kagawa analyzed genes that were transiently expressed along with TROP2 and found OVOL1. Although Kagawa had expected OVOL1 might promote pluripotency, his experiments using the popular gene editing tool CRISPR showed that suppressing OVOL1 did not prevent reprogramming. Rather, cells without OVOL1 grew rapidly and could still be reprogrammed to iPS cells, but at the same time partially reprogrammed cells also flourished.
"OVOL1 suppression increased the number of iPS cells, but also partially reprogrammed cells. We compromised purity for quantity," says Kagawa.
The findings suggest that with more KLF4, cells which choose the alternative identity marked by TROP2 also activate OVOL1, which puts their growth in check. Many genes have been recognized as crucial for starting the reprogramming into iPS cells. Woltjen argues that the discovery of OVOL1 shows how other genes are important to prevent cells from entering alternative pathways that deviate from the final iPS cell destination.
"Reprogramming involves a number of cell fate decisions," says Woltjen. "After more than 10 years of iPS cell technology, there is still much to learn about the process."
Paper Details
- Journal: Stem Cell Reports
- Title: OVOL1 Influences the Determination and Expansion of iPSC Reprogramming Intermediates
- Authors: Harunobu Kagawa1, Ren Shimamoto2, Shin-Il Kim1, Fabian Oceguera-Yanez1, Takuya Yamamoto1, Timm Schroeder2, and Knut Woltjen1,3
- Author Affiliations:
- Department of Life Science Frontiers, Center for iPS Cell Research and Application (CiRA),
Kyoto University, Kyoto, Japan - Department of Biosystems Science and Engineering, ETH Zurich, Basel, Switzerland
- Hakubi Center for Advanced Research, Kyoto University, Kyoto, Japan
- Department of Life Science Frontiers, Center for iPS Cell Research and Application (CiRA),






















