
Research Activities
Research Activities
Publications
October 17, 2023
Delineating the dynamic transcriptional and epigenetic landscape regulating hematopoiesis
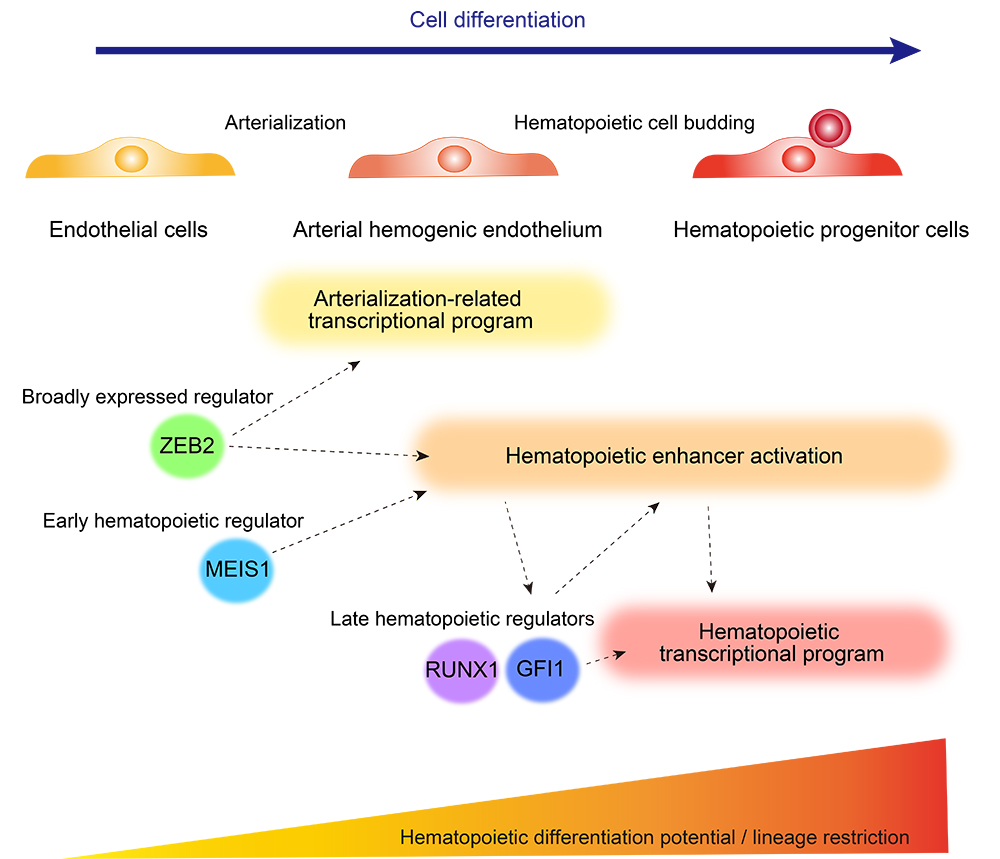
Cell differentiation, the process through which cells attain cellular identity and unique functional characteristics, is primarily driven by cell type-specific enhancers activated by environmental cues or pre-existing regulators from an earlier developmental stage. However, for many cell types, like hematopoietic progenitor cells (HPCs), there is no single master regulator that controls differentiation, so multiple factors must work together intricately to carefully promote stepwise cell type-specific differentiation programs. The precise timing and sequence of enhancer activation are critical for ensuring cells follow the differentiation trajectory properly, but much of the details remain a mystery.
Leveraging a human embryonic stem cell (ESC)-derived hematopoietic cell differentiation system Saito and his team of researchers previously described, they collected ESCs, endothelial cells (an intermediate stage between ESCs and HPCs in the differentiation trajectory), and HPCs at various time points during differentiation to profile their transcriptomes and enhancer signatures by RNA sequencing (RNA-seq) and acetylated-histone chromatin immunoprecipitation and sequencing (H3K27ac ChIP-seq), respectively. RNA-seq recapitulated the gradual transcriptome remodeling of the cells from an ESC-like state to an HPC-like state, enriched with genes involved in lymphoid and myeloid differentiation, thus validating the differentiation system. Conversely, activated enhancers clustered into 3 groups, depending on the timing of activation (group 1, exclusively HPCs; group 2, activated in late-stage endothelial cells before HPC differentiation; and group 3, active mainly in endothelial cells but downregulated in HPCs). Confirmed by current knowledge of several key genes involved in hematopoietic cell differentiation, these data suggest enhancer activation occurs before hematopoietic lineage specification and drives characteristic gene expression.
By examining transcription factor binding motifs in the activated enhancers of each cluster, the researchers reasoned that this represents a perfect opportunity to identify factors involved in hematopoietic enhancer activation. In particular, they focused on enriched motifs activated before HPC differentiation located near hematopoietic cell differentiation-related genes and identified ZEB2 binding motifs to be enriched in cluster-1 and -2 enhancers. Furthermore, ZEB2 expression was detected in the endothelial cell and HPC differentiation stages, precisely matching the transition period during which it would be required. To test the role of ZEB2 in regulating hematopoiesis, the researchers deleted ZEB2 in ESCs and induced pluripotent stem cells (iPSCs) using the CRISPR/Cas9 genome editing system and found that neither (ZEB2 knockout (KO) ESCs nor iPSCs) could produce hematopoietic cells, thus suggesting that ZEB2 is essential to HPC development. Notably, when examining their global gene expression pattern, ZEB2 KO cells on day 10 of HPC differentiation resembled day 7 endothelial cells, thus indicating the progression to HPC has been halted. Furthermore, via H3K27ac ChIP-seq, the research team found a massive decrease of activated enhancers in ZEB2 KO cells, solidifying ZEB2's role in rendering the epigenetic landscape ideal for endothelial cells to differentiate into HPCs during development.
In addition, the researchers further separated cluster 2 enhancers to specifically focus on those with decreased activity in the absence of ZEB2 to identify ZEB2-dependent factors that may also be vital to HPC differentiation. Through this effort, they identified MEIS1 binding motifs to be enriched at enhancers affected by ZEB2 deletion and found it to be a target gene of ZEB2. To better understand the relationship between ZEB2 and MEIS1 in regulating hematopoiesis, the research team deleted MEIS1 separately and observed HPC differentiation to be significantly disrupted. Similar to ZEB2 KO cells, transcriptome analysis revealed an inability of MEIS1 KO cells to activate the hematopoietic transcriptional program. Nonetheless, some MEIS1 KO cells did differentiate into HPCs, albeit at much lower efficiency, and their global gene expression pattern showed a wild-type-like transcriptome profile. Moreover, by deleting both ZEB2 and MEIS1, the researchers found ZEB2/MEIS1 double KO cells to have more pronounced impairment in HPC differentiation, suggesting a partially redundant role for these two proteins to activate the hematopoietic transcriptional program. However, only ZEB2 re-expression could partially rescue defects in hematopoietic enhancer activation, as MEIS1 activation failed to accomplish the same outcome, thereby highlighting the differential requirement for ZEB2 and MEIS2.
Through this study, Saito and his research team identified the independent contributions of ZEB2 and MEIS1 to hematopoiesis. In the process, they elucidated the role of ZEB2, a regulator broadly expressed from endothelial cells, to be crucial for pushing endothelial cells along the developmental trajectory toward becoming HPCs. How ZEB2 and MEIS1 regulate hematopoietic enhancer activation will be vital to improving ex vivo HPC generation from iPSCs for regenerative medicine and drug discovery purposes.
Paper Details
- Journal: iScience
- Title: ZEB2 and MEIS1 independently contribute to hematopoiesis via early hematopoietic enhancer activation
- Authors:
Yohko Kitagawa1,*, Akihiro Ikenaka1, Ryohichi Sugimura1,2, Akira Niwa1, Megumu K. Saito1,*
*:Corresponding authors - Author Affiliations:
- Center for iPS Cell Research and Application, Kyoto University
- Li Ka Shing Faculty of Medicine, University of Hong Kong






















