
Research Activities
Research Activities
Publications
November 17, 2023
Using single-antibodies as a new ingenious tool to build bio-circuitry
For cells to function properly and adapt to changes in the internal and external environment, there is a constant need to sense changes and react to them appropriately. For instance, cells have evolved a wide range of transcriptional (i.e., DNA→RNA), translational (i.e., RNA→protein), and even post-translational (i.e., protein modifications such as phosphorylation, or adding a phosphate group to specific locations on a protein) regulatory mechanisms to detect changes in nutrient and metabolite levels to modulate nutrient uptake, metabolism, and waste metabolite removal properly. Another example is the immune cells in our bodies that constantly try to sense foreign DNA or RNA as signs of invading organisms and turn on certain genes to initiate an appropriate defensive response to potential threats.
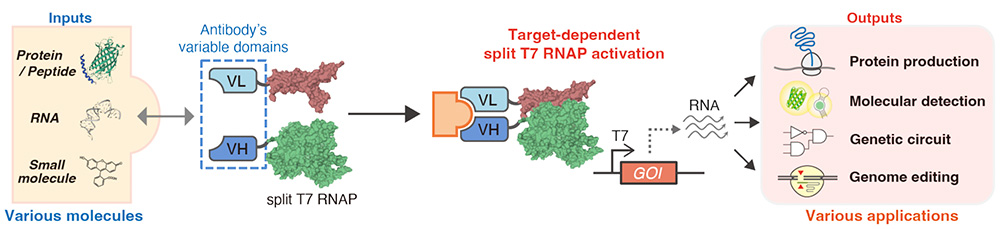
In synthetic biology, scientists have leveraged natural transcriptional and translational regulators to customize biological circuits to control cellular functions in desirable ways. As we enter a new era of bioengineering and regenerative medicine, the demand for novel ways to build bio-circuitry is greater than ever. In a recent study, Saito and his team took a page out of nature's instruction manual and ingeniously applied antibodies, highly evolved proteins capable of binding a broad range of targets such as DNA, RNA, proteins, and essentially any small molecules, to detect specific molecules as inputs and control transgene transcription as outputs.
Specifically, antibodies from most mammals contain a variable region, a three-dimensional structure formed by a heavy and light chain, that binds a certain substrate (target) with high affinity. The researchers recognized that when the target molecule is present inside cells, it will interact separately with the heavy and light chains, effectively bringing them into proximity. They thus attached the variable region (heavy and light chains separately) from antibodies known to interact with specific target molecules to two parts of a split T7 RNA polymerase (RNAP), a protein designed to spontaneously reassemble when brought close together and transcribe RNA from an artificial DNA transgene, to drive expression of reporter genes as an output to signal positive target molecule detection. In the study, the researchers revealed the power and flexibility of their so-called target-dependent RNAP (TdRNAP) system to sense a variety of target molecules, including an antigenic peptide from the yeast transcription factor GCN4, the FLAG peptide, enhanced green fluorescence protein (EGFP), a specific RNA sequence from the hepatitis C virus (HCV), and the fluorescent small molecule, fluorescein, by simply switching between different variable region sequences for target detection. Furthermore, by combining different RNAP variants into the system, they also demonstrated the creation of multilayer biological circuits for either signal amplification or orthogonal signal transduction. Aside from using this TdRNAP system simply for target molecule detection, the research team also demonstrated its application for cell-specific genome editing by using the system to induce guide RNA (gRNA) expression to trigger the CRISPR/Cas9-mediated deletion of a transgene previously incorporated into the genome of an experimental cell line.
The research team believes their new TdRNAP system to be an invaluable new tool for constructing bio-circuity to detect a variety of intracellular molecules and control cell functions. Undoubtedly, this system holds enormous promise for potential applications in improving the efficacy and safety of future gene and cell therapies.
Paper Details
- Journal: Nature Communications
- Title: Target-dependent RNA polymerase as universal platform for gene expression control in response to intracellular molecules
- Authors: Shodai Komatsu1,2, Hirohisa Ohno1, Hirohide Saito1,2*
*:Corresponding authors - Author Affiliations:
- Department of Life Science Frontiers, Center for iPS Cell Research and Application (CiRA),
Kyoto University - Graduate School of Medicine, Kyoto University
- Department of Life Science Frontiers, Center for iPS Cell Research and Application (CiRA),






















