
Research Activities
Research Activities
Publications
July 01, 2025
Splitting up for precise cell type-specific gene regulation
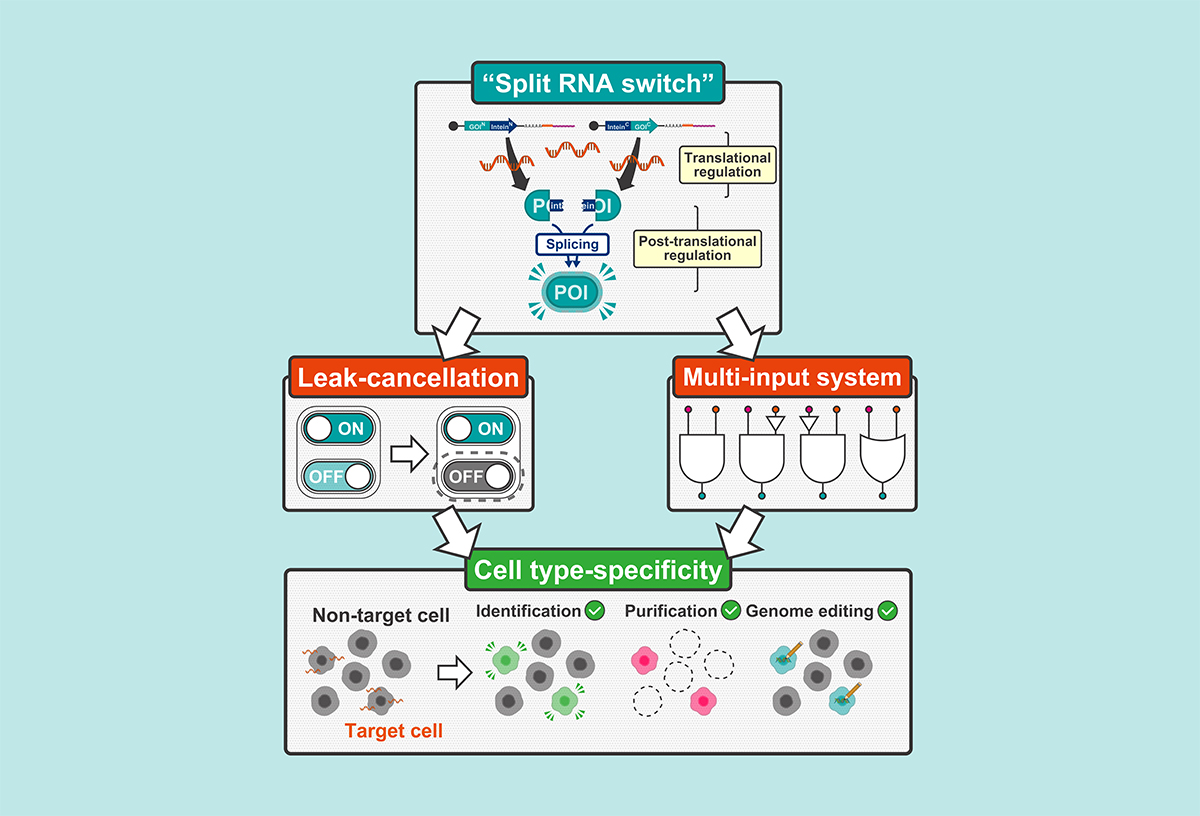
An RNA switch is a synthetic mRNA molecule designed to regulate the translation of a gene in response to specific intracellular signals, such as microRNAs (miRNAs) or proteins. These switches act as programmable molecular logic gates that can turn gene expression on or off depending on the presence of target biomolecules, making them powerful tools for cell type-specific gene regulation. However, conventional RNA switches often suffer from low signal-to-noise ratios due to leaky translation and the difficulty of identifying unique molecular markers for specific cell types.
To overcome these limitations, the research team introduced the split RNA switch, which uses protein splicing to integrate outputs from multiple RNA switches. This system employs split-inteins, protein fragments that rejoin post-translationally to form a functional protein only when both fragments are present. By pairing ON switches that encode the N- and C-terminal fragments of a protein with OFF switches that produce inactive fragments, the system suppresses unintended protein expression in non-target cells. This strategy improved the signal-to-noise ratio of miRNA-responsive switches by more than 25-fold.
The researchers demonstrated the versatility of the split RNA switch across various applications. These included fluorescent reporters for imaging, antibiotic resistance genes for cell selection, and the suicide gene HSV-TK for inducing cell death. In each case, the system enabled cell type-specific control based on endogenous miRNA activity.
The team also applied the split RNA switch to the CRISPR-Cas9 genome editing platform, enabling miRNA-dependent gene editing in human cells, including iPS cells. In a cellular disease model of Duchenne muscular dystrophy, the system achieved over 45-fold specificity in editing the gene for dystrophin, minimizing off-target effects.
To further expand its capabilities, the researchers constructed multi-input logic gates using orthogonal split-inteins. These circuits could detect combinations of miRNAs and proteins, enabling complex intracellular decision-making. A three-input AND gate was successfully demonstrated for the first time using RNA-only components, activating gene expression only when all three target miRNAs were present.
In addition, a toggle-like system using two fluorescent proteins improved the binary separation of cell populations in flow cytometry, enhancing classification accuracy over conventional RNA switches.
This study is the first to combine mRNA-based translational regulation with post-translational protein splicing. The split RNA switch offers a modular, scalable, and RNA-only platform for precise gene control, with strong potential for gene therapy, regenerative medicine, and synthetic biology. This approach opens new avenues for designing "intelligent" RNA therapeutics and lays the groundwork for next-generation treatments capable of adapting to complex cellular environments.
Paper Details
- Journal: Nature Communications
- Title: Split RNA switch orchestrates pre- and post-translational control to enable cell type-specific gene expression
- Authors: Itsuki Abe1,2,3,4, Hirohisa Ohno1*, Megumi Mochizuki1, Karin Hayashi1, Hirohide Saito1,3*
*: Corresponding authors - Author Affiliations:
- Center for iPS Cell Research and Application, Kyoto University
- Graduate School of Medicine, Kyoto University
- Institute for Quantitative Biosciences, The University of Tokyo
- Department of Bioengineering School of Engineering, The University of Tokyo






















