Research Activities
Research Activities
Principal Investigators
Dept. of Life Science Frontiers
Risa K. Kawaguchi (Junior Associate Professor)

Risa K. Kawaguchi Ph.D.
- Lab Website
- Research Progress in FY2024
- Contact: rkawaguchi-g*cira.kyoto-u.ac.jp
- Please change * to @
Research Overview
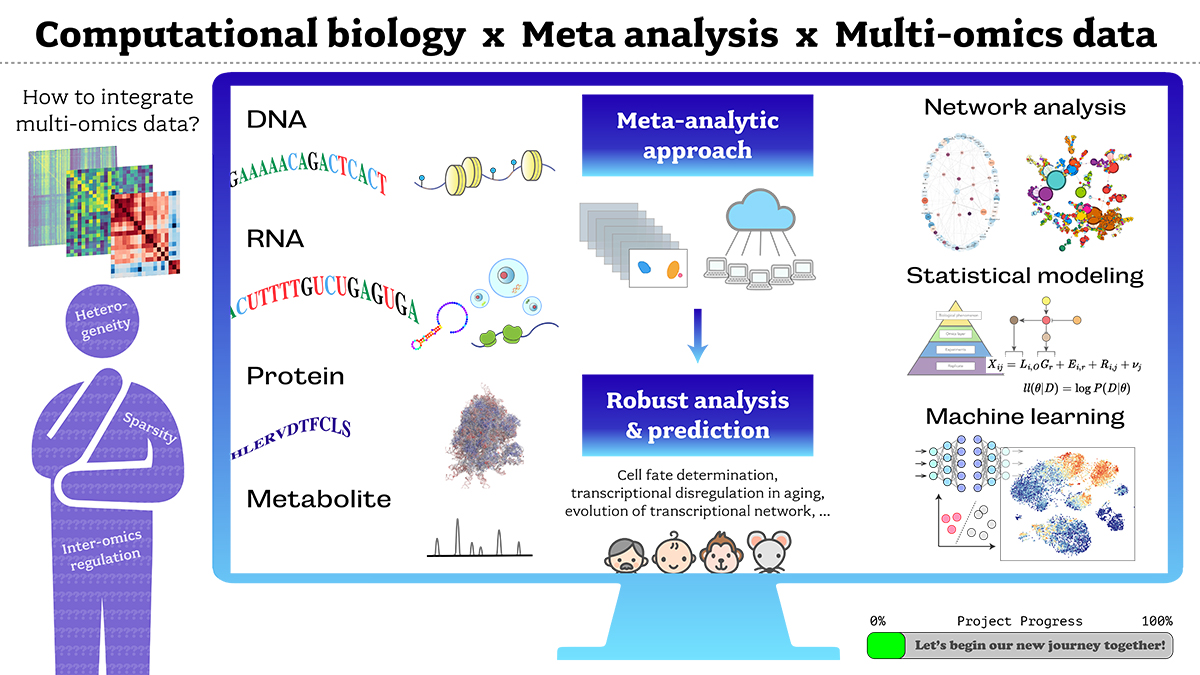
A comprehensive analysis of "omics" data from high-throughput technologies enhances our understanding of how biological regulatory networks produce robust biological systems at a single-cell level. However, the integration of multimodal omics data requires careful statistical modeling to overcome underlying batch effects and systematic biases between different datasets. Further powering this approach, advancements in machine learning have driven significant breakthroughs in our ability to predict previously undetectable traits, such as cell-fate determination, by exploiting the high dimensionality of parameter space.
We aim to develop a novel approach for meta-analytic comparisons of large-scale life science data that integrates technologies and methods from information science, statistics, mathematical modeling, and machine learning. This integrative analysis of omics data will be used to construct a global co-expression network, a powerful resource for linking genetic and transcriptomic perturbations to causal factors such as stochastic variability, aging-dependent network disruption, and changes in robustness and complexity during evolution.
The current research subjects of our laboratory are as follows:
1) Co-expression network analysis using RNA-seq
2) Single-cell RNA-seq and ATAC-seq analysis to discover cell-type specific enhancers
3) RNA structure and post-transcriptional regulation analysis
4) Evolutionary analysis of regulatory sequences
Rather than limiting our analysis to human cell lines and iPS cells, we will apply our approach to a wide range of mammalian species to decipher the universal regulatory mechanism that produces a robust transcriptional profile from the relatively error-prone transcription process.