
Research Activities
Research Activities
Publications
August 28, 2023
Exploring new avenues in DMD treatment: CRISPR-Cas3's multi-exon skipping approach
Duchenne muscular dystrophy (DMD) is a degenerative muscle disease in which genomic mutations in dystrophin, a gene critical for maintaining muscle cells, change the reading frame of the protein (frameshift), resulting in the inability to produce the dystrophin protein. Currently, there is no general curative treatment for all DMD patients.
Exon skipping, which corrects the protein reading frame, has been attracting attention as a means of restoring dystrophin protein production, and nucleic acid drugs that skip exon 53 are now available in Japan and the United States. However, current methods of inducing exon skipping are applicable only to a limited number of mutations (8-13%). A research group at the National Center for Neurology and Psychiatry (NCNP) in Japan targeted exons 45 to 55 using a nucleic acid drug in mice in 2012, with several independent research teams subsequently reporting that a similar multiple exon skipping (MES)-based strategy was effective for more than 60% of DMD patients, thus providing new hope for an increased number of people living with DMD.
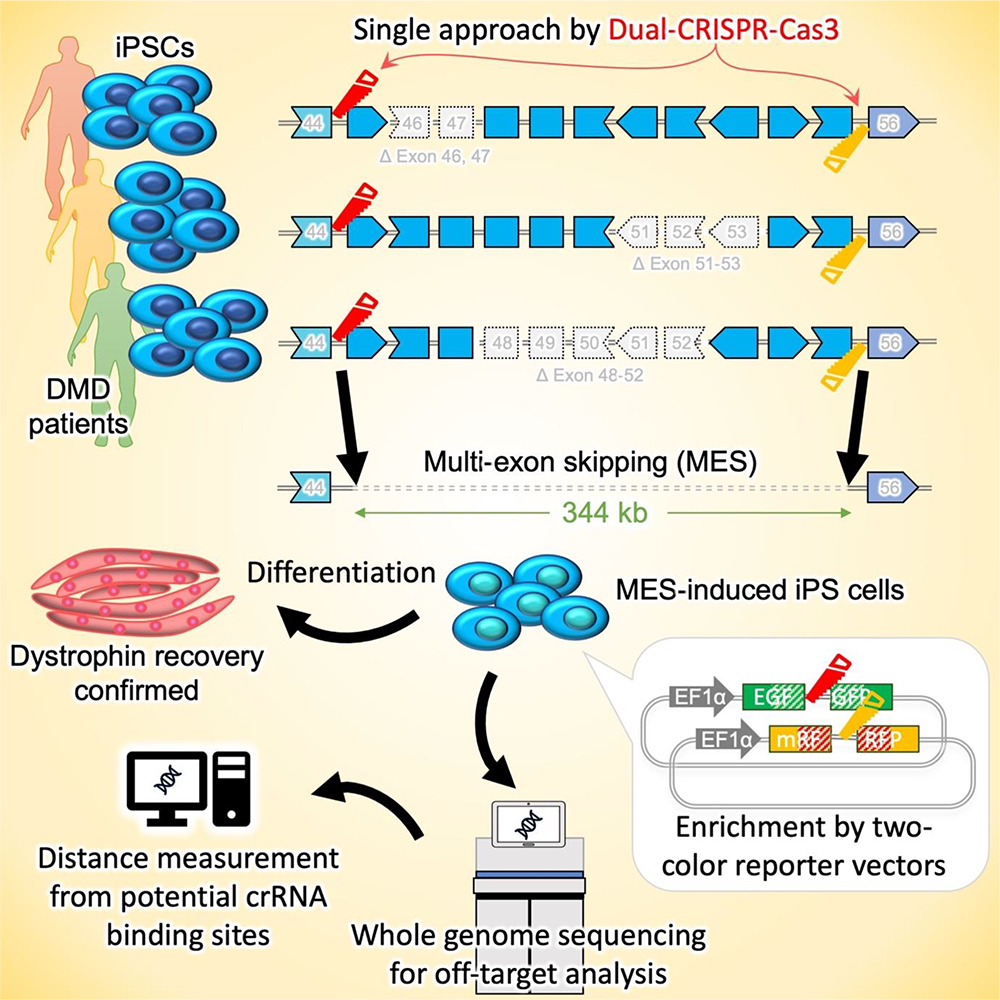
As an alternative therapeutic strategy, several groups have been examining the possibility of inducing exon skipping at the genomic DNA level using CRISPR-Cas9 (CiRA News, November 27, 2014) and CRISPR-Cas3 genome editing technology (CiRA News, December 6, 2019). CRISPR-Cas3 cuts several kb of genome sequence from the targeted point in a unidirectional manner, a characteristic that Associate Professor Hotta and his group hypothesized would be ideal for MES and could be leveraged to treat DMD patients.
In the current study, they developed a dual CRISPR-Cas3 system that could induce a large genomic deletion (340 kb ~) spanning the dystrophin exon 45-55 region. This approach holds promise for a wide range of DMD mutations, thus broadening its applicability. To verify the effectiveness of the dual CRISPR-Cas3 system, the researchers used DMD patient-derived induced pluripotent stem cells (DMD-iPSCs) with three distinct mutations. They also developed a two-color SSA (single-strand annealing)-based reporter system to enrich the population of genome-edited cells, thereby making the process more efficient.
Furthermore, the team conducted whole-genome sequencing and distance analysis to detect potential off-target deletions near the targeted crRNA binding sites and observed no significant off-target effects, supporting the safety and accuracy of the dual CRISPR-Cas3 system.
Overall, this research highlights the potential of the dual CRISPR-Cas3 system as a promising tool to induce large genomic deletions for MES in DMD patients with a wide range of mutation patterns. In addition to DMD, these findings have vast potential to impact other diseases in which a large genomic deletion represents a viable treatment option. By utilizing genome editing technologies like CRISPR-Cas3, there are high hopes to develop long-lasting and effective solutions for patients with DMD or other devastating disorders.
The results of this study were published online in Stem Cell Reports on August 24, 2023.
Paper Details
- Journal: Stem Cell Reports
- Title: Dual CRISPR-Cas3 system for inducing multi-exon skipping in DMD patient-derived iPSCs
- Authors: Yuto Kita1, Yuya Okuzaki2, Youichi Naoe1, Joseph Lee1, Uikyu Bang1, Natsumi Okawa1,
Akane Ichiki1, Tatsuya Jonouchi1, Hidetoshi Sakurai1, Yusuke Kojima1, Akitsu Hotta1,3*
*:Corresponding author - Author Affiliations:
- Center for iPS Cell Research and Application, Kyoto University
- Nagoya University Graduate School of Bioagricultural Sciences, Avian Bioscience Research Center
- Takeda-CiRA Joint Program






















